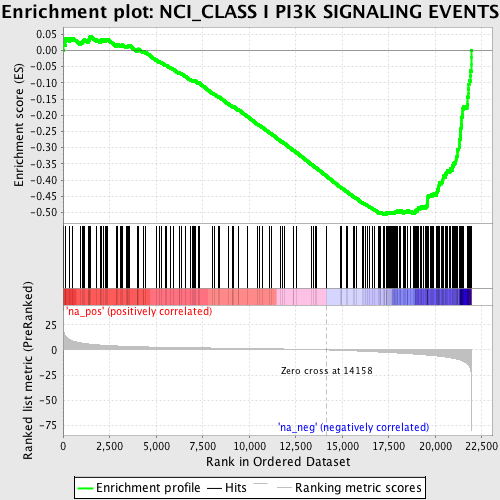
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | NCI_CLASS I PI3K SIGNALING EVENTS |
| Enrichment Score (ES) | -0.5057789 |
| Normalized Enrichment Score (NES) | -2.1713161 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.1802745E-4 |
| FWER p-Value | 0.0040 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL5 | 11 | 22.605 | 0.0193 | No | ||
| 2 | RAN | 122 | 13.946 | 0.0265 | No | ||
| 3 | YWHAQ | 140 | 13.364 | 0.0374 | No | ||
| 4 | CDK4 | 343 | 10.371 | 0.0372 | No | ||
| 5 | HRAS | 510 | 8.966 | 0.0375 | No | ||
| 6 | FCGR2B | 927 | 7.120 | 0.0246 | No | ||
| 7 | PLCG2 | 1020 | 6.821 | 0.0263 | No | ||
| 8 | ARFIP2 | 1070 | 6.660 | 0.0299 | No | ||
| 9 | FGR | 1144 | 6.449 | 0.0322 | No | ||
| 10 | XPO1 | 1342 | 5.926 | 0.0283 | No | ||
| 11 | INPPL1 | 1375 | 5.849 | 0.0320 | No | ||
| 12 | E2F1 | 1414 | 5.788 | 0.0353 | No | ||
| 13 | MAPK1 | 1423 | 5.768 | 0.0400 | No | ||
| 14 | BLK | 1450 | 5.692 | 0.0438 | No | ||
| 15 | KPNB1 | 1789 | 5.095 | 0.0327 | No | ||
| 16 | PPARG | 1999 | 4.807 | 0.0273 | No | ||
| 17 | BAD | 2011 | 4.790 | 0.0310 | No | ||
| 18 | UBE2D3 | 2039 | 4.762 | 0.0339 | No | ||
| 19 | CDKN2A | 2159 | 4.589 | 0.0325 | No | ||
| 20 | CAMK2G | 2267 | 4.454 | 0.0315 | No | ||
| 21 | PTPRK | 2304 | 4.414 | 0.0337 | No | ||
| 22 | NFATC2 | 2387 | 4.322 | 0.0337 | No | ||
| 23 | SH3BP5 | 2857 | 3.902 | 0.0155 | No | ||
| 24 | TP53 | 2880 | 3.885 | 0.0179 | No | ||
| 25 | CDKN1A | 2944 | 3.835 | 0.0184 | No | ||
| 26 | IL3 | 3065 | 3.756 | 0.0162 | No | ||
| 27 | AKT1S1 | 3163 | 3.688 | 0.0149 | No | ||
| 28 | EIF4A1 | 3164 | 3.687 | 0.0182 | No | ||
| 29 | CASP3 | 3387 | 3.501 | 0.0110 | No | ||
| 30 | CASP9 | 3432 | 3.473 | 0.0121 | No | ||
| 31 | FOSL1 | 3479 | 3.447 | 0.0130 | No | ||
| 32 | KRAS | 3504 | 3.433 | 0.0149 | No | ||
| 33 | IL2RA | 3564 | 3.387 | 0.0151 | No | ||
| 34 | EIF4EBP1 | 3974 | 3.161 | -0.0009 | No | ||
| 35 | BTK | 3984 | 3.157 | 0.0014 | No | ||
| 36 | PPP5C | 4007 | 3.142 | 0.0032 | No | ||
| 37 | PPP2R5D | 4047 | 3.115 | 0.0041 | No | ||
| 38 | SLC3A2 | 4297 | 2.996 | -0.0047 | No | ||
| 39 | IBTK | 4328 | 2.985 | -0.0035 | No | ||
| 40 | EIF4G1 | 4441 | 2.936 | -0.0061 | No | ||
| 41 | POU2F2 | 5005 | 2.719 | -0.0296 | No | ||
| 42 | POU2F1 | 5182 | 2.660 | -0.0354 | No | ||
| 43 | MAF | 5280 | 2.628 | -0.0375 | No | ||
| 44 | YWHAG | 5501 | 2.556 | -0.0454 | No | ||
| 45 | INSR | 5561 | 2.539 | -0.0459 | No | ||
| 46 | SOS1 | 5789 | 2.466 | -0.0542 | No | ||
| 47 | RHEB | 5913 | 2.426 | -0.0577 | No | ||
| 48 | PIK3R1 | 6230 | 2.327 | -0.0702 | No | ||
| 49 | IFNG | 6243 | 2.320 | -0.0687 | No | ||
| 50 | EGR4 | 6366 | 2.287 | -0.0723 | No | ||
| 51 | HCK | 6566 | 2.233 | -0.0795 | No | ||
| 52 | KPNA1 | 6863 | 2.150 | -0.0913 | No | ||
| 53 | MAP3K7 | 6975 | 2.124 | -0.0945 | No | ||
| 54 | RAC1 | 7002 | 2.119 | -0.0938 | No | ||
| 55 | CD72 | 7056 | 2.104 | -0.0944 | No | ||
| 56 | NFKB1 | 7091 | 2.093 | -0.0942 | No | ||
| 57 | FASLG | 7259 | 2.047 | -0.1001 | No | ||
| 58 | CLTB | 7302 | 2.037 | -0.1002 | No | ||
| 59 | CD19 | 8029 | 1.852 | -0.1320 | No | ||
| 60 | IRF4 | 8141 | 1.829 | -0.1355 | No | ||
| 61 | CD22 | 8363 | 1.778 | -0.1441 | No | ||
| 62 | RNF128 | 8389 | 1.773 | -0.1437 | No | ||
| 63 | CD79A | 8876 | 1.654 | -0.1646 | No | ||
| 64 | PPP3CA | 9099 | 1.601 | -0.1734 | No | ||
| 65 | SLC2A4 | 9107 | 1.600 | -0.1723 | No | ||
| 66 | YES1 | 9129 | 1.594 | -0.1719 | No | ||
| 67 | PPP3CC | 9402 | 1.530 | -0.1831 | No | ||
| 68 | NFKBIB | 9422 | 1.525 | -0.1826 | No | ||
| 69 | AKAP5 | 9904 | 1.409 | -0.2035 | No | ||
| 70 | CALM1 | 10454 | 1.273 | -0.2277 | No | ||
| 71 | PRKCZ | 10572 | 1.242 | -0.2320 | No | ||
| 72 | PLD2 | 10726 | 1.208 | -0.2379 | No | ||
| 73 | PDE3B | 11073 | 1.116 | -0.2529 | No | ||
| 74 | RPS6 | 11187 | 1.089 | -0.2571 | No | ||
| 75 | BLNK | 11697 | 0.938 | -0.2797 | No | ||
| 76 | DOK1 | 11766 | 0.915 | -0.2821 | No | ||
| 77 | CALM2 | 11871 | 0.882 | -0.2861 | No | ||
| 78 | AP2A1 | 12394 | 0.726 | -0.3094 | No | ||
| 79 | IL4 | 12402 | 0.724 | -0.3091 | No | ||
| 80 | JUN | 12565 | 0.666 | -0.3160 | No | ||
| 81 | TBX21 | 13363 | 0.371 | -0.3524 | No | ||
| 82 | PIM1 | 13451 | 0.338 | -0.3561 | No | ||
| 83 | CSF2 | 13564 | 0.288 | -0.3610 | No | ||
| 84 | RAP1A | 13631 | 0.258 | -0.3638 | No | ||
| 85 | SFN | 14174 | -0.015 | -0.3887 | No | ||
| 86 | REL | 14913 | -0.486 | -0.4222 | No | ||
| 87 | GSK3A | 14949 | -0.501 | -0.4234 | No | ||
| 88 | PTGS2 | 14970 | -0.509 | -0.4239 | No | ||
| 89 | SRC | 15241 | -0.705 | -0.4357 | No | ||
| 90 | CD40LG | 15266 | -0.717 | -0.4362 | No | ||
| 91 | GRB2 | 15610 | -0.952 | -0.4511 | No | ||
| 92 | IRS1 | 15669 | -0.991 | -0.4529 | No | ||
| 93 | MAP2K1 | 15741 | -1.040 | -0.4553 | No | ||
| 94 | TNF | 16079 | -1.307 | -0.4696 | No | ||
| 95 | RIPK2 | 16143 | -1.360 | -0.4713 | No | ||
| 96 | TSC2 | 16253 | -1.451 | -0.4751 | No | ||
| 97 | PRKDC | 16339 | -1.517 | -0.4777 | No | ||
| 98 | ZAP70 | 16444 | -1.608 | -0.4810 | No | ||
| 99 | PRKACA | 16598 | -1.752 | -0.4865 | No | ||
| 100 | SHC1 | 16723 | -1.857 | -0.4906 | No | ||
| 101 | PRKCH | 16959 | -2.083 | -0.4996 | No | ||
| 102 | TRAF6 | 16980 | -2.105 | -0.4987 | No | ||
| 103 | AKT1 | 17066 | -2.193 | -0.5007 | No | ||
| 104 | RHOA | 17075 | -2.199 | -0.4991 | No | ||
| 105 | MAPK9 | 17214 | -2.310 | -0.5034 | No | ||
| 106 | ERC1 | 17266 | -2.356 | -0.5037 | Yes | ||
| 107 | TIAM1 | 17271 | -2.359 | -0.5018 | Yes | ||
| 108 | PTPN6 | 17353 | -2.437 | -0.5034 | Yes | ||
| 109 | CALM3 | 17354 | -2.438 | -0.5013 | Yes | ||
| 110 | MAP3K14 | 17377 | -2.462 | -0.5001 | Yes | ||
| 111 | PRKAB1 | 17420 | -2.502 | -0.4999 | Yes | ||
| 112 | FOXP3 | 17466 | -2.555 | -0.4997 | Yes | ||
| 113 | PLEKHA1 | 17535 | -2.637 | -0.5005 | Yes | ||
| 114 | NUP214 | 17596 | -2.687 | -0.5009 | Yes | ||
| 115 | CHUK | 17643 | -2.730 | -0.5006 | Yes | ||
| 116 | ELK1 | 17690 | -2.773 | -0.5003 | Yes | ||
| 117 | PLEKHA2 | 17744 | -2.825 | -0.5003 | Yes | ||
| 118 | MAPK14 | 17811 | -2.903 | -0.5008 | Yes | ||
| 119 | ARF5 | 17829 | -2.926 | -0.4990 | Yes | ||
| 120 | MAP3K8 | 17860 | -2.954 | -0.4978 | Yes | ||
| 121 | ARHGEF6 | 17905 | -3.006 | -0.4972 | Yes | ||
| 122 | MAPK3 | 17962 | -3.067 | -0.4971 | Yes | ||
| 123 | FKBP1A | 17973 | -3.076 | -0.4948 | Yes | ||
| 124 | FKBP8 | 18055 | -3.159 | -0.4958 | Yes | ||
| 125 | AKT3 | 18133 | -3.248 | -0.4965 | Yes | ||
| 126 | ARF1 | 18135 | -3.249 | -0.4937 | Yes | ||
| 127 | BCL3 | 18305 | -3.430 | -0.4984 | Yes | ||
| 128 | MKNK1 | 18365 | -3.498 | -0.4981 | Yes | ||
| 129 | IKBKG | 18397 | -3.530 | -0.4964 | Yes | ||
| 130 | SYK | 18493 | -3.653 | -0.4976 | Yes | ||
| 131 | PAG1 | 18498 | -3.656 | -0.4946 | Yes | ||
| 132 | PRKCD | 18528 | -3.688 | -0.4927 | Yes | ||
| 133 | YWHAB | 18690 | -3.865 | -0.4967 | Yes | ||
| 134 | ATM | 18851 | -4.077 | -0.5005 | Yes | ||
| 135 | EEF2 | 18879 | -4.125 | -0.4981 | Yes | ||
| 136 | DAPP1 | 18923 | -4.178 | -0.4964 | Yes | ||
| 137 | COPA | 18949 | -4.208 | -0.4939 | Yes | ||
| 138 | PRKCA | 18987 | -4.255 | -0.4918 | Yes | ||
| 139 | ARRB2 | 19063 | -4.371 | -0.4915 | Yes | ||
| 140 | RAF1 | 19074 | -4.383 | -0.4881 | Yes | ||
| 141 | CTLA4 | 19075 | -4.385 | -0.4842 | Yes | ||
| 142 | FOS | 19178 | -4.546 | -0.4849 | Yes | ||
| 143 | PTEN | 19245 | -4.642 | -0.4839 | Yes | ||
| 144 | ARFGAP1 | 19249 | -4.646 | -0.4800 | Yes | ||
| 145 | NR4A1 | 19386 | -4.872 | -0.4819 | Yes | ||
| 146 | NFKB2 | 19492 | -5.020 | -0.4824 | Yes | ||
| 147 | PTPRC | 19517 | -5.062 | -0.4790 | Yes | ||
| 148 | CSNK1A1 | 19558 | -5.138 | -0.4764 | Yes | ||
| 149 | EIF4E | 19561 | -5.139 | -0.4720 | Yes | ||
| 150 | BIRC2 | 19566 | -5.145 | -0.4676 | Yes | ||
| 151 | MDM2 | 19572 | -5.152 | -0.4633 | Yes | ||
| 152 | CD79B | 19598 | -5.186 | -0.4599 | Yes | ||
| 153 | ARF6 | 19599 | -5.188 | -0.4554 | Yes | ||
| 154 | CBLB | 19603 | -5.196 | -0.4510 | Yes | ||
| 155 | MAPKAP1 | 19631 | -5.246 | -0.4476 | Yes | ||
| 156 | IKBKB | 19718 | -5.359 | -0.4469 | Yes | ||
| 157 | CAMK4 | 19772 | -5.438 | -0.4445 | Yes | ||
| 158 | CYLD | 19870 | -5.610 | -0.4441 | Yes | ||
| 159 | YWHAZ | 19925 | -5.718 | -0.4415 | Yes | ||
| 160 | PTPN1 | 20053 | -5.976 | -0.4422 | Yes | ||
| 161 | VAV1 | 20066 | -6.018 | -0.4374 | Yes | ||
| 162 | GOSR2 | 20110 | -6.089 | -0.4341 | Yes | ||
| 163 | GATA3 | 20113 | -6.093 | -0.4288 | Yes | ||
| 164 | AP2M1 | 20168 | -6.215 | -0.4258 | Yes | ||
| 165 | YWHAH | 20180 | -6.225 | -0.4209 | Yes | ||
| 166 | PLCG1 | 20195 | -6.259 | -0.4160 | Yes | ||
| 167 | AKT2 | 20209 | -6.297 | -0.4111 | Yes | ||
| 168 | BCL10 | 20231 | -6.352 | -0.4065 | Yes | ||
| 169 | PPP3CB | 20348 | -6.597 | -0.4061 | Yes | ||
| 170 | TBC1D4 | 20378 | -6.661 | -0.4016 | Yes | ||
| 171 | RELB | 20410 | -6.758 | -0.3971 | Yes | ||
| 172 | ARHGEF7 | 20413 | -6.769 | -0.3912 | Yes | ||
| 173 | VAV2 | 20452 | -6.862 | -0.3869 | Yes | ||
| 174 | PIK3CA | 20532 | -7.036 | -0.3844 | Yes | ||
| 175 | FBXW11 | 20554 | -7.084 | -0.3792 | Yes | ||
| 176 | GBF1 | 20604 | -7.224 | -0.3751 | Yes | ||
| 177 | PRKCE | 20653 | -7.365 | -0.3708 | Yes | ||
| 178 | EP300 | 20781 | -7.751 | -0.3699 | Yes | ||
| 179 | RASA1 | 20833 | -7.927 | -0.3653 | Yes | ||
| 180 | PRKCQ | 20917 | -8.226 | -0.3619 | Yes | ||
| 181 | CSNK2A1 | 20926 | -8.269 | -0.3550 | Yes | ||
| 182 | ETS1 | 20973 | -8.470 | -0.3497 | Yes | ||
| 183 | RELA | 21022 | -8.671 | -0.3443 | Yes | ||
| 184 | GGA3 | 21109 | -8.959 | -0.3404 | Yes | ||
| 185 | ITCH | 21112 | -8.974 | -0.3326 | Yes | ||
| 186 | NRAS | 21157 | -9.167 | -0.3266 | Yes | ||
| 187 | CARD11 | 21186 | -9.295 | -0.3197 | Yes | ||
| 188 | CD4 | 21188 | -9.300 | -0.3116 | Yes | ||
| 189 | MAPK8 | 21208 | -9.408 | -0.3042 | Yes | ||
| 190 | CLTA | 21272 | -9.721 | -0.2986 | Yes | ||
| 191 | CDKN1B | 21294 | -9.870 | -0.2909 | Yes | ||
| 192 | JUNB | 21298 | -9.897 | -0.2824 | Yes | ||
| 193 | FYN | 21301 | -9.911 | -0.2738 | Yes | ||
| 194 | ITK | 21334 | -10.083 | -0.2664 | Yes | ||
| 195 | TSC1 | 21353 | -10.209 | -0.2583 | Yes | ||
| 196 | EGR1 | 21359 | -10.247 | -0.2495 | Yes | ||
| 197 | BCL2 | 21371 | -10.300 | -0.2410 | Yes | ||
| 198 | FRAP1 | 21383 | -10.384 | -0.2324 | Yes | ||
| 199 | CREBBP | 21411 | -10.605 | -0.2243 | Yes | ||
| 200 | CSK | 21416 | -10.654 | -0.2152 | Yes | ||
| 201 | HSP90AA1 | 21420 | -10.699 | -0.2059 | Yes | ||
| 202 | GSK3B | 21455 | -10.935 | -0.1979 | Yes | ||
| 203 | RPS6KB1 | 21479 | -11.098 | -0.1892 | Yes | ||
| 204 | TNFAIP3 | 21482 | -11.148 | -0.1795 | Yes | ||
| 205 | NFATC1 | 21534 | -11.557 | -0.1717 | Yes | ||
| 206 | TNFRSF1A | 21703 | -13.816 | -0.1674 | Yes | ||
| 207 | MEF2D | 21742 | -14.522 | -0.1564 | Yes | ||
| 208 | NFATC3 | 21743 | -14.538 | -0.1436 | Yes | ||
| 209 | NFKBIA | 21775 | -15.114 | -0.1318 | Yes | ||
| 210 | LCK | 21778 | -15.166 | -0.1186 | Yes | ||
| 211 | MAP3K1 | 21803 | -15.769 | -0.1059 | Yes | ||
| 212 | YWHAE | 21814 | -16.134 | -0.0922 | Yes | ||
| 213 | MALT1 | 21871 | -18.066 | -0.0789 | Yes | ||
| 214 | PDPK1 | 21879 | -18.696 | -0.0628 | Yes | ||
| 215 | DGKA | 21928 | -24.490 | -0.0436 | Yes | ||
| 216 | EGR3 | 21930 | -24.862 | -0.0218 | Yes | ||
| 217 | EGR2 | 21933 | -25.685 | 0.0006 | Yes |